TF Binding
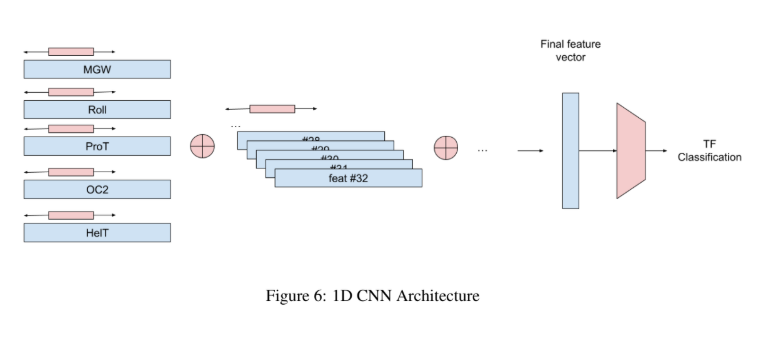
For my graduate course, COMP 561, introduction to bioinformatics, we had a final project where we were tasked with trying to predict the binding sites of transcription factors based on open chromatin region data (ChIP-seq), using physical features to improve prediction. For my project, we created an extensible framework that can test many different Machine Learning models (both classical and deep learning-based), enabling users to compare which architecture works best at predicting transcription factors. We concluded that this classification task remains difficult if motif scores are overlapping (i.e. high motif scores), and likely requires sequence data as well.
More details are available in my report, found here:
and my GitHub found: https://github.com/ehuan2/tf-binding.